

Indian Journal of Science and Technology
DOI: 10.17485/IJST/v13i32.1251
Year: 2020, Volume: 13, Issue: 32, Pages: 3230-3236
Original Article
James Paul T. Madigal1*, Jayson F. Cariaga2, Thiara Celine E. Suarez2,Shirley C. Agrupis1
1Professors, Department of Biological Sciences, College of Arts and Sciences, Mariano Marcos State University, City of Batac, Ilocos Norte, 2906, Philippines. Tel.: (+63)995-4702-204
2Researchers, Research Directorate, Mariano Marcos State University, City of Batac, Ilocos Norte, 2906, Philippines
*Corresponding Author
Tel: (+63)995-4702-204
Email: [email protected]
Received Date:27 July 2020, Accepted Date:09 August 2020, Published Date:31 August 2020
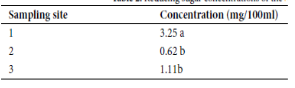
Background/Objectives: Sap from Nypa fruticans are used as feedstock for a bioethanol national program to establish a sustainable and productive village-scale bioethanol industry. The study aimed to establish the physicochemical, microbiological, and molecular characteristics of nipa sap. Methods/Statistical Analysis: Nipa sap samples were collected from three sampling sites at different fermentation time. Physico-chemical parameters such as pH, titratable acidity, and total soluble solids were determined and statistically analyzed using One-way Analysis of Variance and mean ± SD values. Molecular approaches were used to profile and identify yeast species naturally residing in the sap during fermentation and assess the genetic diversity of nipa palm in selected sampling locations. Findings: Sampling site and fermentation time significantly affect the levels of pH, titratable acids, and total soluble solids in the sap samples. Sugar concentration in nipa sap is affected by sampling site: as the sampling site nears the river, the amount of sugars become significantly greater. Molecular studies revealed that among the 12 isolates obtained, five of them (Y24S1, Y48S1, Y72S1, Y24S2, and Y24S3) showed 100% similarity to strain of Pichia occidentalis, three (Y48S2, Y48S3, and Y72S3) were similar (100%) to a strain of Saccharomyces cerevisiae (YPDW9), and the other two isolates (Y72S1 and Y48S3A) were identical to Naganishia randhawae strain HBUMO6885 and Tremellales sp. LM523 respectively with a 100% similarity. PCR amplification of their trnL–trnF cpDNA genome, BLAST and phylogenetic analyses reveals that all nipa plant samples were similar to Nypa fruticans. Applications: The generated information on the effect of sampling site on sugar content of nipa sap samples could be good input in selecting expansion sites for the establishment of nipa plantations. Furthermore, the identified yeast strains can be used as inoculant or starter culture to improve fermentation.
Keywords: Physico-chemical parameter; yeasts molecular profiling; nipa sap fermentation; phylogenetic analysis; genetic diversity
© 2020 James Paul T. Madigal et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee).
Subscribe now for latest articles and news.