

Indian Journal of Science and Technology
Year: 2021, Volume: 14, Issue: 36, Pages: 2806-2814
Original Article
Shazia Irfan1, Shajahan Shabbir Ahmed Rana2, Imran Ali Sani2, Liu Haun2, Mu Weixue3, Ruksana Jabeen1, Nazeer Ahmed2*
1Department of Botany, Faculty of life Sciences, Sardar Bahadur Khan Women’s University, Quetta, Pakistan
2Department of Biotechnology, (Balochistan University of Information Technology, Engineering and Management Sciences), Quetta, Pakistan
3China National Gene Bank, Shenzhen, China
*Corresponding Author
Email: [email protected]
Received Date:08 February 2021, Accepted Date:11 June 2021, Published Date:11 February 2021
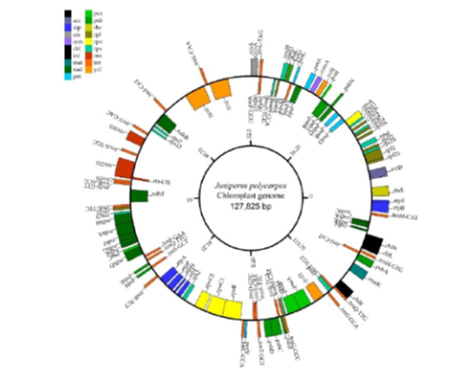
Objectives: To broaden the genetic information base of Juniperus and resolve phylogeny of Juniperus polycarpos through sequencing and characterization of its chloroplast genome. Methods: The chloroplast (cp) genome of J. polycarpos was sequenced and assembled using the Next-Generation Sequencing pairedend reads platform of BGISEQ-500 and annotated using CpGAVAS. The phylogenetic analysis was performed in MEGA7. Findings: Here, we report the complete cp genome sequence of J. polycarpos. The cp genome size is 127,825 bp with a typical circular structure and lack canonical inverted repeats having a total of 119 genes comprised of 82 protein-coding genes, 33 tRNA genes and four rRNA genes. The cp genome encodes 105 single copy genes and five duplicated genes (ndhK, ccsA, rps12, trnE-TTC and trnQ-TTG), and one tetraplicated gene(trnM-CAT). In these genes, 9 genes (rpl2, ycf2, trnA-TGC, trnETTC,rpoC, rpoB, ndhB, ndhA and atpF) harboring a single intron, three genes (accD, rrn23s and ycf3) having two introns and one gene (ycf1) harboring three introns. The overall GC content of J. polycarpos chloroplast DNA was 35%. Phylogenetic analysis among 14 species of order Coniferales based on cp genomes indicated a close relationship between J. polycarpos, J. cedrus and J. communis. Novelty and application: This is the first report on the cp genome of J. polycarpos. The current study is expected to add to the already available genomic resources needed for more comprehensive population genetics studies and resolving phylogenetic relationships of order Coniferales. Besides, it will provide baseline data for future research on Juniperus of Pakistan in particular.
Keywords: BGISeq-500; Cloroplast Genome; Persian Juniper; Phylogeny
© 2021 Irfan et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.