

Indian Journal of Science and Technology
Year: 2020, Volume: 13, Issue: 43, Pages: 4434-4445
Original Article
Dashwa Langbang1, Rahul Dhodapkar2*, KC Premarajan3
1PhD scholar, Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry, 605006, India
2Additional Professor, Department of Microbiology, Jawaharlal Institute of Postgraduate
Medical Education and Research (JIPMER), Puducherry, 605006, India
3Professor, Department of Community Medicine, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry, 605006, India
*Corresponding Author
Email: [email protected]
Received Date:01 February 2020, Accepted Date:17 November 2020, Published Date:08 December 2020
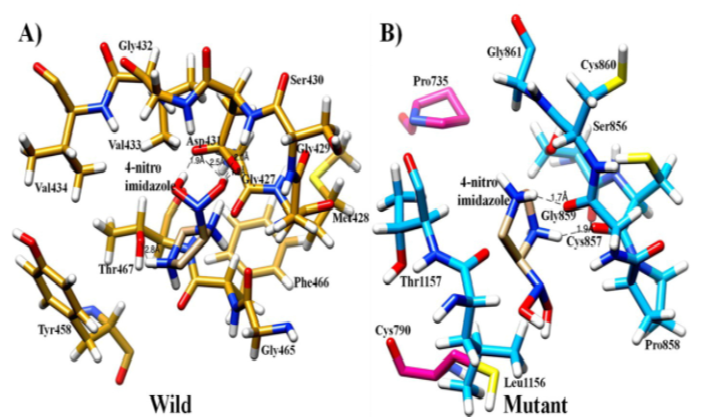
Background/Objective. Giardia intestinalis is a flagella protozoan residing in human intestine causing diarrhea that affecting most common among the children. Usually metronidazole and nitroimidazole were used for the treatment of giardiasis worldwide. Pyruvate Ferredoxin oxidoreductase (PFeO) protease enzyme may play a role inactivation of these drugs that may lead to resistance. Studies regarding drug resistance are very less. Therefore, present study was carried out to predict the structure and characterize them structurally as well as functionally by using appropriate in-silico methods. Methods: The proteins sequences of wild type and mutant strain i.e. Pyruvate Ferredoxin oxidoreductase (PFeO) and Pyruvate flavodoxin oxidoreductase (PFIO) were retracted from the NCBI and aligned using ClustalW. Bioinformatic tools were carried out like molecular dynamics simulations and docking to understand the three-dimensional (3D) conformational structure and interaction behavior of mutant and wild types with the ligand. Findings: The wild complex maintained an approximate 4H-bonds during simulation while the mutant complex could hardly maintain single H-bond by the end of the simulation, suggesting that 4-nitroimidazole is highly stable in the wild complex. Since the compound occupies this binding site by making stable H-bond with key interacting residues, the function of mutant may be affected. For the very reason, the mutant may be resistant to the nitroimidazole. Applications: Molecular dynamic simulation and docking results have increased our understanding of resistance mechanisms and may also help for the design derivatives of nitroimidazole that inhibit protein function more efficiently.
Keywords: Molecular dynamic simulation; Nitroimidazole resistance; Pyruvate Ferredoxin Oxidoreductase (PFeO) mutant type; Pyruvate Flavodoxin Oxidoreductase (PFIO) wild type; Giardia intestinalis
© 2020 Langbang et al.This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.