

Indian Journal of Science and Technology
Year: 2020, Volume: 13, Issue: 27, Pages: 2747-2754
Original Article
Nazir Ahmed1*, Muhammad Ali1, Hidayatullah2, Zahid Mustafa1, Tabeel Tariq Bashir1, Farooq Shehzad1
1Center for Advanced Studies in Vaccinology & Biotechnology (CASVAB), University of Balochistan, Pakistan
2Agriculture Research Institute (ARI), Quetta, Pakistan
Corresponding author
Email: [email protected]
Received Date:31 March 2020, Accepted Date:18 May 2020, Published Date:31 July 2020
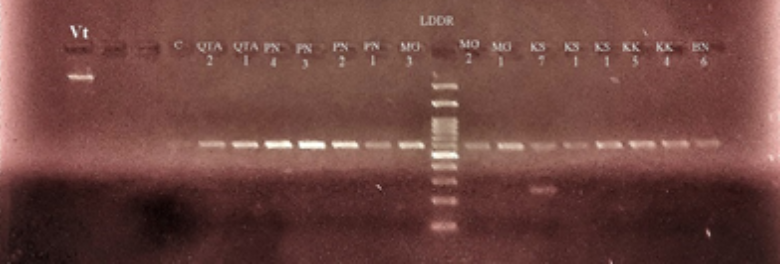
Background/objectives: Orobanche is one of the important parasites of solanaceae crops and causes heavy yield losses. This study aimed to use DNA barcoding technique for identification of Orobanche species. Methods: Surveillance study was conducted in the year 2018 on tomato crops grown in four different districts in Balochistan. From these tomato fields, 15 Orobanche plant samples were collected and put in polyethylene bags containing silica gel for drying. The dried samples were processed for DNA extraction using CTAB method. The extracted DNA was confirmed through gel electrophoresis which were further amplified on PCR using rbcL, matK primers. Then, the PCR products were processed for DNA sequencing using Sanger method. Further, the DNA sequences were edited and aligned with BioEdit and MegaX software and the aligned sequences were matched on NCBI for BLAST. The FASTA results of both partial and/or complete genome were run on MegaX for phylogenetic analysis. Findings: The success rates for PCR amplification was 100% for rbcl primer, while it was 68% for matK primer. However, the resolution power of matK was higher than that obtained from rbcL as manifested in the rate of DNA acquisition sequence. The DNA barcode results of 15 samples revealed 99.16% rate of DNA acquisition sequence for identification of P. ramosa and 96.41 for P. purpurea. While, Rbcl rate of DNA sequencing show no results. Application/Improvement: from this study it was inferred that matK gene performed better than rbcL in Orobanche species identification and proved as a potential gene for other pseudogene plants species identification.
Keywords: Orobanche; species; DNA barcode; rbcL; matK; tomato
© 2020 Ahmed, Ali, Mustafa, Bashir, Shehzad. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee).
Subscribe now for latest articles and news.