

Indian Journal of Science and Technology
Year: 2023, Volume: 16, Issue: 35, Pages: 2835-2844
Original Article
Khushbu Agrawal1*, Tarun Patel2, Shavi Thakur1, Kruti Patel1, Sumit Mittal3
1Department of Chemistry, Shree Maneklal M. Patel Institute of Sciences and Research, Sector 15/23, Kadi Sarva Vishwavidyalaya, Gandhinagar, 382023, Gujarat, India
2Department of Chemistry, Sir P T Science College, HNGU, Modasa
3School of Advanced Sciences and Languages (SASL), VIT Bhopal University, Kothri Kalan, Near Indore Road, Bhopal, 466114, Madhya Pradesh, India
*Corresponding Author
Email: [email protected]
Received Date:21 February 2023, Accepted Date:08 August 2023, Published Date:19 September 2023
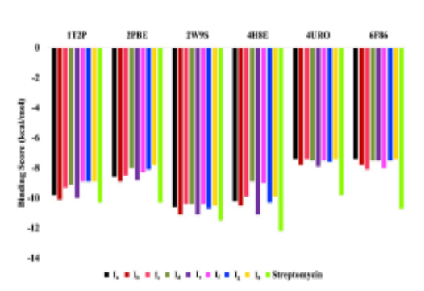
Objective: To explore the possibility of obtaining effective and harmless antibacterial drugs by conventional synthesis of new piperidine analogs. Methods: The target compound was carried out by cyclization reaction involving aromatic aldehydes and amine and further evaluated for their antimicrobiological assay using agar disc diffusion method. Computational investigation such as density functional theory (DFT) was carried out by B3LYP/6-31G (d,p) method to evaluate their electronic characteristics. Absorption, Distribution, Metabolism, and Excretion (ADME) properties were evaluated using the Swiss ADME server. The Antimicrobial activity was investigated using molecular docking and that molecule was prepared using MGL Tools 1.5.7. Auto Dock Vina 1.2.0. Findings: Among all the synthesized compounds 5a to 5h, compounds 5b and 5e showed good anti-bacterial properties against three pathogenic bacterial strains. B. subtilis is inhibited by compound 5b at a concentration of 26 mm. B. subtilis is inhibited by compound 5e at a concentration of 25 mm. Molecular Docking revealed that molecules 5b and 5e consistently showed a large binding affinity for all six target proteins, which was similar to that of the reference molecule, streptomycin. Drug-likeness predictions and the ADME analysis showed that most of the molecules do not violate any of the five Lipinski rule while some have only one or two violations, primarily because of high molecular weight. Novelty: In order to investigate the anti-bacterial impact of piperidine derivatives, a lot of work has been put into their synthesis and evaluation. These results might influence the creation and advancement of anti-microbial medication candidates that are more potent.
Keywords: Piperidine; Antimicrobial; SAR Study; Molecular Docking; ADME Study; DFT
© 2023 Agrawal et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.