

Indian Journal of Science and Technology
DOI: 10.17485/IJST/v17i27.1110
Year: 2024, Volume: 17, Issue: 27, Pages: 2873-2879
Original Article
S S Machchhar1∗, U L Solanki1, H S Sanghavi1
1Assistant Professor, Computer Engineering Department, Government Engineering College, Bhavnagar, Gujarat, India
*Corresponding Author
Email: [email protected]
Received Date:05 April 2024, Accepted Date:29 June 2024, Published Date:19 July 2024
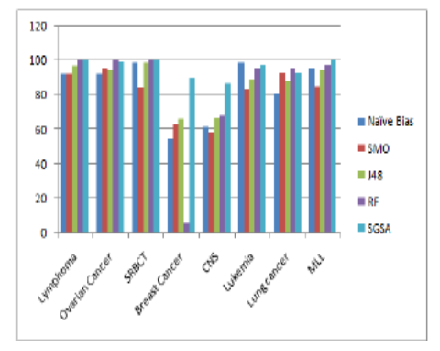
Objectives: To make a study on hybrid scheme of selecting a most representative subset of genes to improve microarray data classification accuracy using machine learning algorithm. Method: ML-based SGSA approach proposed here has two parts of execution on gene expression datasets. First it utilizes an entropy based IGKullback–Leibler divergence to select the most informative genes. Subsequently, an attribute evaluation performed using correlation-based feature selection. After that a random forest based classifier is employed with 10-fold cross-validation. The proposed method involves data pre processing, testing, training, fitting an algorithm and then finds the best accuracy with comparable CPU cost. Findings: The rationale behind this study is that only most informative genes are submitted to classifier for classification task. Proven accuracy by this approach is 98.48 for Lymphoma, 89.69 for Breast, 86.67 for CNS, 97.22 for Leukemia, 98.4 for Lung cancer, 96.6 for MLL, 97.21 for Ovarian cancer and 98.5 for SRBCT over traditional machine learning algorithms like naïve bias, J48 and SMO. These results demonstrate the effectiveness of the suggested approach in accurately classifying tumors. The numerical illustration also showed that the new estimator is more efficient in terms of CPU cost. Novelty: The major problem with traditional machine learning algorithms is that all features are treated equally important during the classification process which makes them susceptible to the influence of outliers and difficult to find a meaningful class in the dataset. In this study, classification accuracy is improved by processing the most informative features in the classifiers. The primary contribution of the research is a hybrid ML model which uses IG based feature selection followed by correlation analysis. The result is then fed to RF based classifier which significantly enhance accuracy as well as CPU cost.
Keywords: Machine Learning, Classification, Correlation, Feature Selection, Gene Expression
© 2024 Machchhar et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.