

Indian Journal of Science and Technology
DOI: 10.17485/IJST/v16i14.2163
Year: 2023, Volume: 16, Issue: 14, Pages: 1082-1089
Original Article
Agnik Haldar1, Ajay Kumar Singh1*
1Department of Bioinformatics, Center for Biological Sciences (Bioinformatics), Central University of South Bihar, Panchanpur Road,Tekari, Gaya, Fathehpur, 824236, India
*Corresponding Author
Email: [email protected]
Received Date:13 November 2022, Accepted Date:25 January 2023, Published Date:12 April 2023
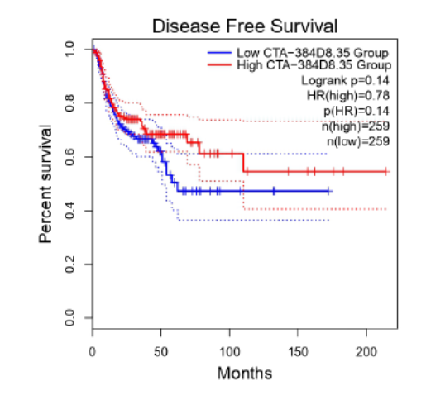
Objective: Gingivobuccal cancer is a subtype of oral cancer, prevalent in developing countries where the use of tobacco and Areca (betel) nut is rampant. This study aims to search through the publically available NCBI data to find comparisons between the normal and cancer-affected transcripts of RNA, which are molecules that copied the genetic information from DNA and that will be “read” as instructions to produce other molecules in the body. Using bioinformatic tools, we analyzed the variations in the transcripts and examined how they were correlated with other molecular processes in the body. We specifically investigated the role of “long noncoding RNAs (lncRNAs)” because it is speculated that they play an active role in the causality of cancer. Our analysis found a correlation of CTA-384D8.35 lncRNA with gingivobuccal cancer after analyzing the Hazard Ratio using the Cox Proportional-Hazard Model and is expected to pave the way for future studies. Methods: In this study, transcriptomic analysis was performed on a publicly available gingivobuccal cancer dataset obtained from NCBI. We developed a novel method for identification of annotated lncRNAs, which helped us select and correlate the lncRNA transcripts with the gingivobuccal cancer disease. Furthermore, the results were validated using the GEPIA2 tool for integrated analysis of co-lncRNA and GEPIA databases. Findings: From 24 RNA-Seq samples (12 normal samples and 12 tumor samples), a total of 16,065 potential lncRNA transcripts were recognized. From these 16,065 lncRNA transcripts, 1201 lncRNA transcripts were further filtered out based on a threshold adjusted p value of <0.01. We were able to for the first time identify these 1201 lncRNA transcripts, which were not annotated for gingivobuccal squamous cell carcinoma previously. Integrated analysis of co-lncRNA and GEPIA databases revealed that a total of 11 lncRNAs were upregulated, whereas 29 lncRNAs were downregulated. Furthermore, CTA-384D8.35 showed a significant correlation with disease-free survival time in HNSC. CTA-384D8.35 which is downregulated in gingivobuccal cancer has been correlated with tumor stage, metastasis, cancer status, clinical stage, and grade. Novelty: In this report we are for the first time correlating the CTA-384D8.35 lncRNA with the gingivobuccal cancer disease with the help of statistical and survival analysis. Furthermore,1201 lncRNA transcripts, which have previously not been annotated for gingivobuccal squamous cell carcinoma, were identified in this study based on a threshold adjusted p value of <0.01. Finally we have also introduced a novel method of identifying annotated lncRNAs in a disease as well as improved upon the NGS data analysis pipeline by determining the strandness of the raw RNASeq data. Differential expression of lncRNAs could possibly play a significant role in carcinogenesis. Therefore the identification of lncRNAs gains priority, which are speculated to play a role in the mechanism and formation of various forms of cancer. For identification and analysis of lncRNAs, a comparison of their expression in normal and affected tissues is likely to confirm their involvement in the disease or the lack thereof. The introduction of a novel methodology along with the identification of CTA-384D8.35 lncRNA paves the way for further analysis pertaining to lncRNAs as prognostic or diagnostic biomarkers. Since CTA-384D8.35 lncRNA has previously been correlated with a number of cancer hallmarks, it gains significant traction to be considered as an important aspect in the biomarker identification pertaining to gingivobuccal cancer. This study has reported for the first time the lncRNAs that are prevalent in gingivobuccal squamous cell carcinoma.
Keywords: Gingivobuccal cancer; lncRNA; oral cancer; epigenetics; NGS data analysis
© 2023 Haldar & Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.