

Indian Journal of Science and Technology
Year: 2022, Volume: 15, Issue: 3, Pages: 127-135
Original Article
Usha Yadav1, Sandeep Yadav1, Dinesh C Sharma1*
1School of Life Sciences, Starex University, Gurugram, India (122413) *
*Corresponding Author
Email: [email protected]
Received Date:01 October 2021, Accepted Date:13 January 2021, Published Date:04 February 2022
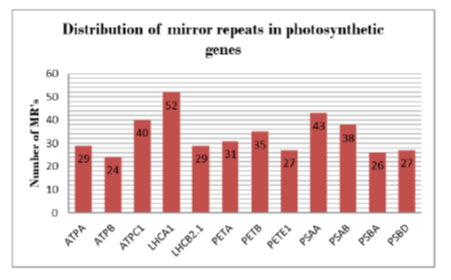
Objectives: The underlying work explores mirror sequences in the photosynthetic genes of Arabidopsis thaliana. At present, these sequences are standing at the forefront to be explored for their origin, distribution and function in plants. Methods: FPCB, a recently developed bioinformatics approach was utilized for identification of mirror sequences. It is a three step strategy based on pattern matching of alignments, produced after aligning gene sequence and its complement using mega-BLAST. This algorithm was quick and efficient enough to characterize a range of mirror sequences. Findings: All the analyzed genes were reported to harbor great variety of mirror sequences at quite high frequencies. LHCA1 gene have the highest total count of these sequences and ATPB gene have lowest of all. A total of 401 unique mirror sequences of different lengths and compositions were reported in the twelve selected genes. Promoter motifs were found to be greatly enriched with these repeats. Eleven mirror sequences of significant lengths were also reported using the above approach. Novelty: This work is the very first attempt to characterize photosynthetic genes of Arabidopsis thaliana for mirror repeats. This will further aggravate efforts to develop fingerprinting techniques based on these unique mirror sequences, which are very powerful tools to study taxonomic and evolutionary relationships. Mirror sequences are also potential candidates as drug delivery systems and in molecular medicine.
Keywords: Mirror repeats; HDNA; FPCB; photosynthetic genes; Arabidopsis thaliana
© 2022 Yadav et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.