

Indian Journal of Science and Technology
Year: 2024, Volume: 17, Issue: 11, Pages: 1078-1086
Original Article
Ruhina Karani1*, Jay Mehta1, Jay Mistry1, Harit Koladia1, Chetashri Bhadane1
1Department of Computer Engineering, Dwarkadas J. Sanghvi College of Engineering, Mumbai, Maharashtra, India
*Corresponding Author
Email: [email protected]
Received Date:12 January 2024, Accepted Date:12 February 2024, Published Date:07 March 2024
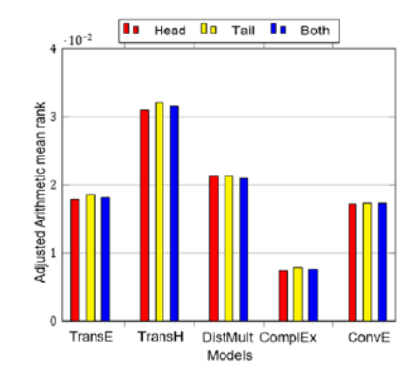
Objective: This research aims to enhance genetic counselors' efficiency in analyzing genetic data across diverse medical settings, spanning prenatal scans, tumor testing, carrier typing, and Fluorescence In Situ Hybridization (FISH), etc. The objective is to employ graph-based techniques for identifying potential gene-disease associations and recommending personalized medical interventions. The application scope extends to areas like personalized medicine, newborn screening, and genetic probing. Methods: The study utilizes a novel technique within the PrimeKG genetic graph database, predicting gene-disease associations. Ensembles are constructed from six models - TransE, TransD, TransR, TransH, ComplEx, and DistMult. The hits@10 metric evaluates the model's effectiveness, measuring the accuracy of predictions within the top 10 ranked associations. The ensemble achieves a score of 0.52, indicating a significant proportion of correct predictions in the top ten associations. Findings: The research presents an efficient approach to identify gene-disease associations, demonstrating a high hits@10 metric accuracy (0.52). This method significantly aids medical professionals in making informed patient care decisions, with potential applications in prenatal scans, tumor testing, carrier typing, and more. The findings underscore the utility of graph-based techniques in transforming disease identification and treatment through genetic data analysis. Novelty: This study introduces a unique approach, leveraging ensembles from diverse models, to predict gene-disease associations within the PrimeKG genetic graph database. The hits@10 metric underscores the model's efficiency, presenting a novel and valuable contribution to the field of genetic data analysis and healthcare decision-making.
Keywords: Gene Disease Prediction, Machine Learning, Knowledge Graph Completion, Drug Discovery, Healthcare
© 2024 Karani et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.