

Indian Journal of Science and Technology
Year: 2021, Volume: 14, Issue: 17, Pages: 1398-1405
Original Article
Received Date:28 January 2021, Accepted Date:01 May 2021, Published Date:15 May 2021
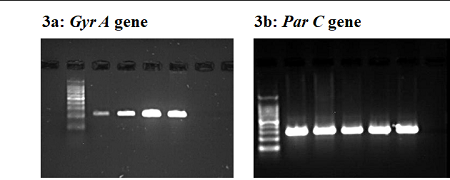
Background and Objective: Gyr A and Par C genes, are known to cause resistance especially in Alterations in proteins of fluoroquinolones. Here, we investigated human pathogens Escherichia coli causing Urinary Tract Infection [UTI] to explore the possible link between the abundance of mutations, and the exposure to fluoroquinolones. In this study, we investigated the occurrence of Gyr A and Par C gene producers among Quinolones resistant [QR] Escherichia Coli isolated Methods: 148 Urine samples were collected from a patients with UT infections. Phenotypically, Ciprofloxacin resistance was screened by micro broth dilution method. Multiplex PCR was carried out to determine the mutations in Gyr A and Par C genes. We have determined partial sequences of the Gyr A and Par C genes of E. coli including the regions analogous to the quinolone resistance-determining region of the E. coli Gyr A gene. Results: Out of 148 urine samples, 100 E. coli were isolated and identified. We analysed 20 quinolone-resistant strains for alterations in Gyr A and Par C. Of these, 11 Gyr A-positive isolates were identified using the Gyr A specific primers and were clearly Ciprofloxacin resistant. The other 9 Ciprofloxacin-resistant isolates were found to have Par C genes using specific primers. We observed an unexpectedly high prevalence of Gyr A than Par C in patients attending a tertiary care hospital by PCR with an estimation of 9.0% (95% confidence interval). This study demonstrated that the number of mutations in QRs of Gyr A and/or Par C was significantly associated with the MICs of quinolones (P<0.01). Conclusion: The Gyr A and Par C genes were detected predominantly in E. coli. The data emerging out of this study helps in understanding the dynamics of this infection and provide inputs for antibiotic policy in the treatment of urinary tract infections.
Keywords: Ciprofloxacin, resistance, Escherichia coli, Polymerase chain reaction, microdilution method
© 2021 Jagathy & Malathi. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Published By Indian Society for Education and Environment (iSee)
Subscribe now for latest articles and news.